Hi, I'm Pratibha, PhD
I'm a Bioinformatician.
I'm passionate about RNA, technology development, biotech, business, and art.
Contact MeAbout Me
Background & InterestsI’m a scientist, bioinformatician, and biotech enthusiast with a background that spans academic research, venture capital, and technology development. My work centers on decoding RNA biology using next-generation sequencing, high-throughput screening, and machine learning—driven by a passion for transforming complex data into actionable insights. Beyond science, I’m deeply connected to the arts. Trained in Indian classical music and dance for over 15 years, I now enjoy social dancing, jazz, and curating the perfect R&B playlist.
Skills
My technical levelComputational Biology & Bioinformatics
Expertise in the construction and implementation of analysis pipelines and integrative analysis.- Transcriptome assembly and quantification
- Alternative splicing and polyadenylation analysis
- Long-read and short-read sequencing analysis (bulk and single-cell)
- RNA-protein and Protein-Protein interaction and network analysis
- Multi-omics integration
- Statistical testing and enrichment analysis
Tools, Programming, & Automation
Proficient in widely used programming languages, workflow tools, and high-performance computing resources.- Python, R, Jupyter
- Linux, Bash
- Data visualization
- GitHub
- HPC Cluster Computing
- Snakemake
- AWS
AI / Machine Learning
Expertise in applying AI/ML to biological data and therapeutic discovery.- Deep learning (standard Python packages)
- Predictive modeling
- Feature engineering for biological sequence
- HPC Cluster Computing
- Model interpretation and validation
Data Generation & Experimental Design
Proficient in experimental techniques and design.- High-throughput screening
- Molecular biology techniques
- Cell culture
- Library preparation
Leadership, Communication, & Strategy
Expertise in effective project management, leadership, and communication across cross-disciplinary teams.- Scientific due dilligence
- Project planning
- Public speaking
- Collaboration
Research
Publications, Topics, and Themes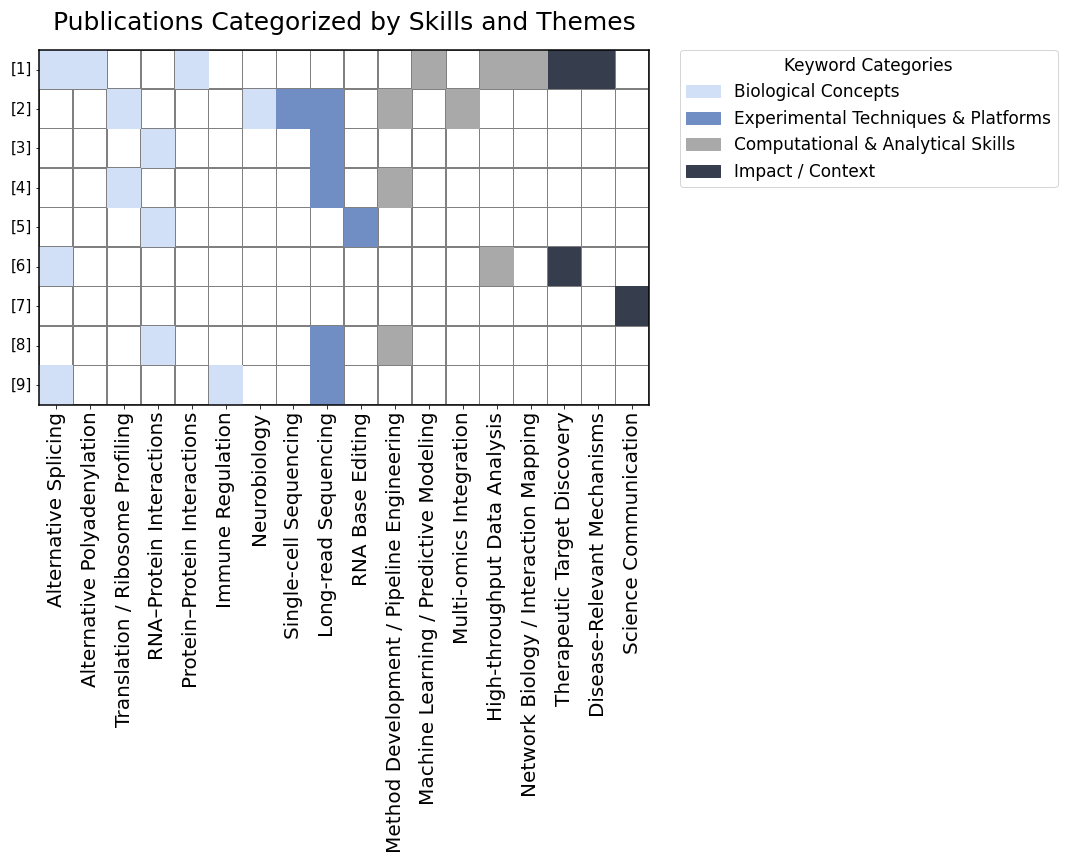
- Jagannatha, P., Yoon, Y., Landry, S., Naritomi, J.T., Zhan, L., Olson, S., Wei, X., Street, L. A., Jovanovic, M., Graveley, B. R., Yeo, G.W., Large-scale discovery of RNA-binding proteins that directly modulate poly(A) site selection (in preparation)
- Zampa, F., Sison, L. S., Kofman, E. R., Choi, S. Y., Jagannatha, P., Nguyen, G. G., Naritomi, J. T., Shin, A., Khorgade, A., Jin, W., Chen, C., Sievert, D. M., Mukhopadhyay, S., Mizrahi, O., Blue, S. M., Marina, R. J., Yang, D., Wang, C. C., Pang, Z., Brannan, K. W., Ye, Li., Stowers, L., Al’ Khafaji, A. M., Lippi, G., Yeo, G. W. Single cell and isoform-specific translational profiling of the mouse brain. (submitted)
- Liang, Q., Yu, T., Kofman, E., Jagannatha, P., Rhine, K., Yee, B. A., Corbett, K. D., Yeo, G. W.. High-sensitivity in situ capture of endogenous RNA-protein interactions in fixed cells and primary tissues. Nat Commun 15, 7067 (2024). https://doi.org/10.1038/s41467-024-50363-4
- Jagannatha, P., Tankka, A. T., Lorenz, D. A., Yu, T., Yee, B. A., Brannan, K. W., Zhou, C. J., Underwood, J. G., & Yeo, G. W. (2024). Long-read Ribo-STAMP simultaneously measures transcription and translation with isoform resolution. Genome research, gr.279176.124. Advance online publication. https://doi.org/10.1101/gr.279176.124
- Medina-Munoz, H. C., Kofman, E., Jagannatha, P., Boyle, E. A., Yu, T., Jones, K. L., Mueller, J. R., Lykins, G. D., Doudna, A. T., Park, S. S., Blue, S. M., Ranzau, B. L., Kohli, R. M., Komor, A. C., & Yeo, G. W. (2024). Expanded palette of RNA base editors for comprehensive RBP-RNA interactome studies. Nature communications, 15(1), 875. https://doi.org/10.1038/s41467-024-45009-4
- Schmok, J. C., Jain, M., Street, L. A., Tankka, A. T., Schafer, D., Her, H. L., Elmsaouri, S., Gosztyla, M. L., Boyle, E. A., Jagannatha, P., Luo, E. C., Kwon, E. J., Jovanovic, M., & Yeo, G. W. (2024). Large-scale evaluation of the ability of RNA-binding proteins to activate exon inclusion. Nature biotechnology, 10.1038/s41587-023-02014-0. Advance online publication. https://doi.org/10.1038/s41587-023-02014-0
- Boyle, E. A., Goldberg, G., Schmok, J. C., Burgado, J., Izidro Layng, F., Grunwald, H. A., Balotin, K. M., Cuoco, M. S., Chang, K. C., Ecklu-Mensah, G., Arakaki, A. K. S., Ahmed, N., Garcia Arceo, X., Jagannatha, P., Pekar, J., Iyer, M., DASL Alliance, & Yeo, G. W. (2023). Junior scientists spotlight social bonds in seminars for diversity, equity, and inclusion in STEM. PloS one, 18(11), e0293322. https://doi.org/10.1371/journal.pone.0293322
- Brannan K. W., Chaim I. A., Marina R. J., Yee B. A., Kofman E. R., Lorenz D. A., Jagannatha, P., Dong K. D.,Madrigal A. A., Underwood J. G. & Yeo G. W. (2021). Robust single-cell discovery of RNA targets of RNA-binding proteins and ribosomes. Nat Methods 18, 507–519. https://doi.org/10.1038/s41592-021-01128-0
- Robinson, E. K.*, Jagannatha, P.*, Covarrubias, S., Cattle, M., Smaliy, V., Safavi, R., Shapleigh, B., Abu-Shumays, R., Jain, M., Cloonan, S. M., Akeson, M., Brooks, A. N., & Carpenter, S. (2021). Inflammation drives alternative first exon usage to regulate immune genes including a novel iron-regulated isoform of Aim2. eLife, 10, e69431. https://doi.org/10.7554/eLife.69431